Efficient Synthesis of Peptide and Protein Functionalized Pyrrole-Imidazole Polyamides Using Native Chemical Ligation
Abstract
:1. Introduction

2. Results
2.1. Synthesis of Cysteine-Functionalized Pyrrole-Imidazole (Py–Im) Polyamide


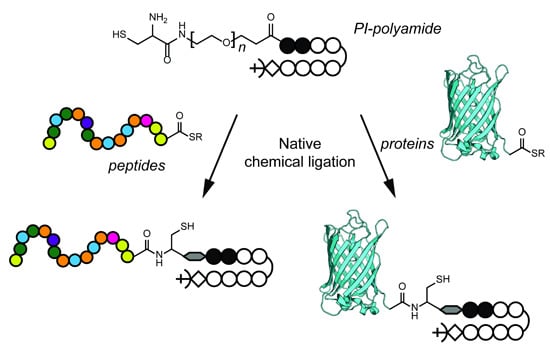
2.2. Native Chemical Ligation of Thioester Peptides and Proteins to Cys-Py–Im-Polyamides

2.3. Effect of Conjugation on DNA Binding Properties


3. Discussion and Conclusion
4. Experimental Section
4.1. Fmoc-Mediated Solid Phase Synthesis of the Non- and Tert-BuSH Protected Cysteine Py–Im Polyamide
4.2. Fmoc-Mediated Solid Phase Peptide Synthesis of Nbz-Peptide
4.3. Protein Expression and Purification
4.4. Native Chemical Ligation Reactions
4.5. Surface Plasmon Resonance
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zhang, F.; Nangreave, J.; Liu, Y.; Yan, H. Structural DNA Nanotechnology: State of the art and future perspective. J. Am. Chem. Soc. 2014, 136, 11198–11211. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Liu, Y.; Yan, H. Designer DNA nanoarchitectures. Biochemistry 2009, 48, 1663–1674. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Pal, S.; Nangreave, J.; Deng, Z.; Liu, Y.; Yan, H. DNA origami with complex curvatures in three-dimensional space. Science 2011, 332, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, C.M. Semisynthetic DNA-protein conjugates for biosensing and nanofabrication. Angew. Chem. Int. Ed. 2010, 49, 1200–1216. [Google Scholar] [CrossRef] [PubMed]
- Saccà, B.; Niemeyer, C.M. Functionalization of DNA nanostructures with proteins. Chem. Soc. Rev. 2011, 40, 5910–5921. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.; Giselbrecht, S.; Rapp, B.E.; Hirtz, M.; Niemeyer, C.M. Advances in DNA-directed immobilization. Curr. Opin. Chem. Biol. 2014, 18, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Liu, M.; Liu, Y.; Yan, H. Spatially-interactive biomolecular networks organized by nucleic acid nanostructures. Acc. Chem. Res. 2012, 45, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Sano, T.; Smith, C.L.; Cantor, C.R. Immuno-PCR: Very sensitive antigen detection by means of specific antibody-DNA conjugates. Science 1992, 258, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, C.M.; Sano, T.; Smith, C.L.; Cantor, C.R. Oligonucleotide-directed self-assembly of proteins: Semisynthetic DNA—Streptavidin hybrid molecules as connectors for the generation of macroscopic arrays and the construction of supramolecular bioconjugates. Nucleic Acids Res. 1994, 22, 5530–5539. [Google Scholar] [CrossRef] [PubMed]
- Weizmann, Y.; Braunschweig, A.B.; Wilner, O.I.; Cheglakov, Z.; Willner, I. Supramolecular aptamer–thrombin linear and branched nanostructures. Chem. Commun. 2008, 4888–4890. [Google Scholar] [CrossRef] [PubMed]
- Cheglakov, Z.; Weizmann, Y.; Braunschweig, A.B.; Wilner, O.I.; Willner, I. Increasing the complexity of periodic protein nanostructures by the rolling-circle-amplified synthesis of aptamers. Angew. Chem. Int. Ed. 2008, 47, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Nakata, E.; Liew, F.F.; Uwatoko, C.; Kiyonaka, S.; Mori, Y.; Katsuda, Y.; Endo, M.; Sugiyama, H.; Morii, T. Zinc-finger proteins for site-specific protein positioning on DNA-origami structures. Angew. Chem. Int. Ed. 2012, 51, 2421–2424. [Google Scholar] [CrossRef] [PubMed]
- Furman, J.L.; Badran, A.H.; Ajulo, O.; Porter, J.R.; Stains, C.I.; Segal, D.J.; Ghosh, I. Toward a general approach for RNA-templated hierarchical assembly of split-proteins. J. Am. Chem. Soc. 2010, 132, 11692–11701. [Google Scholar] [CrossRef] [PubMed]
- Prokup, A.; Deiters, A. Interfacing synthetic DNA logic operations with protein outputs. Angew. Chem. Int. Ed. 2014, 53, 13192–13195. [Google Scholar] [CrossRef] [PubMed]
- Pazos, E.; Mosquera, J.; Vázquez, M.E.; Mascareñas, J.L. DNA recognition by synthetic constructs. ChemBioChem 2011, 12, 1958–1973. [Google Scholar] [CrossRef] [PubMed]
- Dervan, P.B.; Doss, R.M.; Marques, M.A. Programmable DNA binding oligomers for control of transcription. Curr. Med. Chem. Anti-Cancer Agents 2005, 5, 373–387. [Google Scholar] [CrossRef]
- Trauger, J.W.; Baird, E.E.; Dervan, P.B. Recognition of 16 base pairs in the minor groove of DNA by a pyrrole−imidazole polyamide dimer. J. Am. Chem. Soc. 1998, 120, 3534–3535. [Google Scholar] [CrossRef]
- Warren, C.L.; Kratochvil, N.C.S.; Hauschild, K.E.; Foister, S.; Brezinski, M.L.; Dervan, P.B.; Phillips, G.N.; Ansari, A.Z. Defining the sequence-recognition profile of DNA-binding molecules. Proc. Natl. Acad. Sci. USA 2006, 103, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Puckett, J.W.; Muzikar, K.A.; Tietjen, J.; Warren, C.L.; Ansari, A.Z.; Dervan, P.B. Quantitative microarray profiling of DNA-binding molecules. J. Am. Chem. Soc. 2007, 129, 12310–12319. [Google Scholar] [CrossRef] [PubMed]
- Gottesfeld, J.M.; Neely, L.; Trauger, J.W.; Baird, E.E.; Dervan, P.B. Regulation of gene expression by small molecules. Nature 1997, 387, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, L.A.; Gulizia, R.J.; Trauger, J.W.; Baird, E.E.; Mosier, D.E.; Gottesfeld, J.M.; Dervan, P.B. Inhibition of RNA polymerase II transcription in human cells by synthetic DNA-binding ligands. Proc. Natl. Acad. Sci. USA 1998, 95, 12890–12895. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, A.; Noguchi, K.; Minoshima, M.; Kashiwazaki, G.; Kanda, T.; Katayama, K.; Mitsuhashi, J.; Bando, T.; Sugiyama, H.; Sugimoto, Y. DNA ligand designed to antagonize EBNA1 represses Epstein–Barr virus-induced immortalization. Cancer Sci. 2011, 102, 2221–2230. [Google Scholar] [CrossRef] [PubMed]
- Mapp, A.K.; Ansari, A.Z.; Ptashne, M.; Dervan, P.B. Activation of gene expression by small molecule transcription factors. Proc. Natl. Acad. Sci. USA 2000, 97, 3930–3935. [Google Scholar] [CrossRef] [PubMed]
- Arndt, H.-D.; Hauschild, K.E.; Sullivan, D.P.; Lake, K.; Dervan, P.B.; Ansari, A.Z. Toward artificial developmental regulators. J. Am. Chem. Soc. 2003, 125, 13322–13323. [Google Scholar] [CrossRef] [PubMed]
- Stafford, R.L.; Dervan, P.B. The reach of linear protein-DNA dimerizers. J. Am. Chem. Soc. 2007, 129, 14026–14033. [Google Scholar] [CrossRef] [PubMed]
- Stafford, R.L.; Arndt, H.-D.; Brezinski, M.L.; Ansari, A.Z.; Dervan, P.B. Minimization of a protein-DNA dimerizer. J. Am. Chem. Soc. 2007, 129, 2660–2668. [Google Scholar] [CrossRef] [PubMed]
- Krpetić, Ž.; Singh, I.; Su, W.; Guerrini, L.; Faulds, K.; Burley, G.A.; Graham, D. Directed assembly of DNA-functionalized gold nanoparticles using pyrrole–imidazole polyamides. J. Am. Chem. Soc. 2012, 134, 8356–8359. [Google Scholar]
- Patel, S.; Jung, D.; Yin, P.T.; Carlton, P.; Yamamoto, M.; Bando, T.; Sugiyama, H.; Lee, K.-B. NanoScript: A nanoparticle-based artificial transcription factor for effective gene regulation. ACS Nano 2014, 8, 8959–8967. [Google Scholar] [CrossRef] [PubMed]
- Singh, I.; Wendeln, C.; Clark, A.W.; Cooper, J.M.; Ravoo, B.J.; Burley, G.A. Sequence-selective detection of double-stranded DNA sequences using pyrrole–imidazole polyamide microarrays. J. Am. Chem. Soc. 2013, 135, 3449–3457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, T.L.; Nandi, C.K.; Rasched, G.; Parui, P.P.; Brutschy, B.; Famulok, M.; Heckel, A. Polyamide struts for DNA architectures. Angew. Chem. Int. Ed. 2007, 46, 4382–4384. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.L.; Heckel, A. Construction of a structurally defined double-stranded DNA catenane. Nano Lett. 2011, 11, 1739–1742. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Schuster, M.; Bagshaw, C.R.; Rant, U.; Burley, G.A. Site-specific assembly of DNA-based photonic wires by using programmable polyamides. Angew. Chem. Int. Ed. 2011, 50, 2712–2715. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Sadowski, J.P.; Dervan, P.B. Addressing single molecules on DNA nanostructures. Angew. Chem. Int. Ed. 2007, 46, 7956–7959. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Sadowski, J.P.; Dervan, P.B. Programming multiple protein patterns on a single DNA nanostructure. J. Am. Chem. Soc. 2008, 130, 402–403. [Google Scholar] [CrossRef] [PubMed]
- Yoshidome, T.; Endo, M.; Kashiwazaki, G.; Hidaka, K.; Bando, T.; Sugiyama, H. Sequence-selective single-molecule alkylation with a pyrrole–imidazole polyamide visualized in a DNA nanoscaffold. J. Am. Chem. Soc. 2012, 134, 4654–4660. [Google Scholar] [CrossRef] [PubMed]
- Best, T.P.; Edelson, B.S.; Nickols, N.G.; Dervan, P.B. Nuclear localization of pyrrole–imidazole polyamide-fluorescein conjugates in cell culture. Proc. Natl. Acad. Sci. USA 2003, 100, 12063–12068. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.-W.; Tsunaka, Y.; Yokota, H.; Matsumoto, T.; Kashiwazaki, G.; Morinaga, H.; Hashiya, K.; Bando, T.; Sugiyama, H.; Harada, Y. Construction and characterization of Cy3- or Cy5-conjugated hairpin pyrrole–imidazole polyamides binding to DNA in the nucleosome. Biomater. Sci. 2014, 2, 297. [Google Scholar] [CrossRef]
- Jiménez-Balsa, A.; Dodero, V.I.; Mascareñas, J.L. Toward encoding reactivity using double-stranded DNA. Sequence-dependent native chemical ligation of DNA binding polyamides. Tetrahedron 2013, 69, 7847–7853. [Google Scholar] [CrossRef]
- Mapp, A.K.; Dervan, P.B. Preparation of thioesters for the ligation of peptides with non-native substrates. Tetrahedron 2000, 41, 9451–9454. [Google Scholar] [CrossRef]
- Wurtz, N.R.; Turner, J.M.; Baird, E.E.; Dervan, P.B. Fmoc solid phase synthesis of polyamides containing pyrrole and imidazole amino acids. Org. Lett. 2001, 3, 1201–1203. [Google Scholar] [CrossRef] [PubMed]
- Neely, L.; Trauger, J.W.; Baird, E.E.; Dervan, P.B.; Gottesfeld, J.M. Importance of minor groove binding zinc fingers within the transcription factor IIIA-DNA complex. J. Mol. Biol. 1997, 274, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.J.; Cash, K.J.; Plaxco, K.W. Excimer-based peptide beacons: A convenient experimental approach for monitoring polypeptide-protein and polypeptide-oligonucleotide interactions. J. Am. Chem. Soc. 2006, 128, 14018–14019. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.J.; Cash, K.J.; Hugenberg, V.; Plaxco, K.W. Peptide beacons: A new design for polypeptide-based optical biosensors. Bioconjug. Chem. 2007, 18, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Canosa, J.B.; Dawson, P.E. An efficient fmoc-spps approach for the generation of thioester peptide precursors for use in native chemical ligation. Angew. Chem. Int. Ed. 2008, 120, 6957–6961. [Google Scholar] [CrossRef]
- Johnson, E.C.B.; Kent, S.B.H. Insights into the mechanism and catalysis of the native chemical ligation reaction. J. Am. Chem. Soc. 2006, 128, 6640–6646. [Google Scholar] [CrossRef] [PubMed]
- Kukolka, F.; Müller, B.K.; Paternoster, S.; Arndt, A.; Niemeyer, C.M.; Bräuchle, C.; Lamb, D.C. A single-molecule Förster resonance energy transfer analysis of fluorescent DNA–protein conjugates for nanobiotechnology. Small 2006, 2, 1083–1089. [Google Scholar] [CrossRef] [PubMed]
- Kukolka, F.; Schoeps, O.; Woggon, U.; Niemeyer, C.M. DNA-directed assembly of supramolecular fluorescent protein energy transfer systems. Bioconjug. Chem. 2007, 18, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Gholami, Z.; Brunsveld, L.; Hanley, Q. PNA-induced assembly of fluorescent proteins using DNA as a framework. Bioconjug. Chem. 2013, 24, 1378–1386. [Google Scholar] [CrossRef] [PubMed]
- Reulen, S.W.A.; Dankers, P.Y.W.; Bomans, P.H.H.; Meijer, E.W.; Merkx, M. Collagen targeting using protein-functionalized micelles: The strength of multiple weak interactions. J. Am. Chem. Soc. 2009, 131, 7304–7312. [Google Scholar] [CrossRef] [PubMed]
- Reulen, S.W.A.; Brusselaars, W.W.T.; Langereis, S.; Mulder, W.J.M.; Breurken, M.; Merkx, M. Protein-liposome conjugates using cysteine-lipids and native chemical ligation. Bioconjug. Chem. 2007, 18, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Breurken, M.; Lempens, E.H.M.; Temming, R.P.; Helms, B.A.; Meijer, E.W.; Merkx, M. Collagen targeting using multivalent protein-functionalized dendrimers. Bioorg. Med. Chem. 2011, 19, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.C.C.; Dervan, P.B. Sequence-specific trapping of topoisomerase I by DNA binding polyamide−camptothecin conjugates. J. Am. Chem. Soc. 2001, 123, 8657–8661. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wilson, W.D. Quantitative analysis of small molecule–nucleic acid interactions with a biosensor surface and surface plasmon resonance detection. Methods Mol. Biol. 2010, 613, 1–23. [Google Scholar] [PubMed]
- Hsu, C.F.; Phillips, J.W.; Trauger, J.W.; Farkas, M.E.; Belitsky, J.M.; Heckel, A.; Olenyuk, B.Z.; Puckett, J.W.; Wang, C.C.C.; Dervan, P.B. Completion of a programmable DNA-binding small molecule library. Tetrahedron 2007, 63, 6146–6151. [Google Scholar] [CrossRef] [PubMed]
- Herman, D.M.; Baird, E.E.; Dervan, P.B. Tandem hairpin motif for recognition in the minor groove of DNA by pyrrole–imidazole polyamides. Chem. Eur. J. 1999, 5, 975–983. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janssen, B.M.G.; Van Ommeren, S.P.F.I.; Merkx, M. Efficient Synthesis of Peptide and Protein Functionalized Pyrrole-Imidazole Polyamides Using Native Chemical Ligation. Int. J. Mol. Sci. 2015, 16, 12631-12647. https://doi.org/10.3390/ijms160612631
Janssen BMG, Van Ommeren SPFI, Merkx M. Efficient Synthesis of Peptide and Protein Functionalized Pyrrole-Imidazole Polyamides Using Native Chemical Ligation. International Journal of Molecular Sciences. 2015; 16(6):12631-12647. https://doi.org/10.3390/ijms160612631
Chicago/Turabian StyleJanssen, Brian M. G., Sven P. F. I. Van Ommeren, and Maarten Merkx. 2015. "Efficient Synthesis of Peptide and Protein Functionalized Pyrrole-Imidazole Polyamides Using Native Chemical Ligation" International Journal of Molecular Sciences 16, no. 6: 12631-12647. https://doi.org/10.3390/ijms160612631